Articles
- Page Path
- HOME > Epidemiol Health > Volume 42; 2020 > Article
-
Original Article
Major enteropathogens in humans, domestic animals, and environmental soil samples from the same locality: prevalence and transmission considerations in coastal Odisha, India -
Arpit Kumar Shrivastava1
 , Nirmal Kumar Mohakud2
, Nirmal Kumar Mohakud2 , Swagatika Panda1
, Swagatika Panda1 , Saumya Darshana Patra1
, Saumya Darshana Patra1 , Subrat Kumar1
, Subrat Kumar1 , Priyadarshi Soumyaranjan Sahu1,3
, Priyadarshi Soumyaranjan Sahu1,3
-
Epidemiol Health 2020;42:e2020034.
DOI: https://doi.org/10.4178/epih.e2020034
Published online: May 26, 2020
1Infection Biology Laboratory, School of Biotechnology, Kalinga Institute of Industrial Technology (KIIT) Deemed to be University, Bhubaneswar, India
2Kalinga Institute of Medical Sciences, Kalinga Institute of Industrial Technology (KIIT) University, Bhubaneswar, India
3Department of Microbiology and Immunology, Medical University of the Americas, Nevis, West Indies
- Correspondence: Priyadarshi Soumyaranjan Sahu Department of Microbiology and Immunology, Medical University of the Americas, P.O. Box 701 Charlestown, Nevis, West Indies E-mail: priyadarshi_sahu@yahoo.com
©2020, Korean Society of Epidemiology
This is an open-access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
OBJECTIVES
- Regions with limited sanitation facilities have higher rates of infections with various enteric pathogens. It is therefore important to identify different hosts and their relative contribution to pathogen shedding into the environment, and to assess the subsequent health risks to humans.
-
METHODS
- In this study, human faecal (n=310), animal faecal (n=150), and environmental (soil) samples (n=40) were collected from the same locality and screened for selected enteric pathogens by immunochromatography and/or polymerase chain reaction.
-
RESULTS
- At least 1 microbial agent was detected in 49.0%, 44.7%, and 40.0% of the samples from human, animals, and soil, respectively. Among humans, rotavirus was predominantly detected (17.4%) followed by enteropathogenic Escherichia coli (EPEC) (15.4%), Shigella (13.8), and Shiga toxin-producing E. coli (STEC) (9.7%). Among animals, STEC was detected most frequently (28.0%), and EPEC was the major enteric pathogen detected in soil (30.0%). The detection rate of rotavirus was higher among younger children (≤2 years) than among older children. Single infections were more commonly detected than multiple infections in humans (p<0.01), unlike the observations in animal and soil samples. For diarrhoeagenic E. coli and Shigella, most of the human and animal isolates showed close relatedness, suggesting possible cross-infection between humans and domesticated animals in the area studied.
-
CONCLUSIONS
- The present study provides an improved understanding of the distribution of major enteric pathogens coexisting in humans and animals in the region, thereby suggesting a high potential for possible transmission among livestock and communities residing in the studied locality.
- The burden of gastrointestinal infections remains a major problem, especially in low-income countries, as worldwide data show that infectious diarrhoeal disorders alone account for nearly 0.8 million deaths in children less than 5 years of age annually [1]. Multiple aetiological agents, including bacteria, viruses, and parasites, contribute to the diarrhoea of infective aetiology in humans and animals [2]. These diarrhoeal agents are most commonly transmitted either through contaminated food and water or through the faecal-oral route. Therefore, lack of access to clean water, insufficient hygiene, and inadequate sanitation conditions in resource-poor settings put the community at high-risk of suffering from diarrhoea [3]. The anthroponotic and zoonotic transmission of diarrhoeal diseases occurs through a wide range of environmental reservoirs contaminated with various diarrhoeagenic pathogens common to both humans and animals [4]. In particular, domestication of livestock and pet practices often contribute to the zoonotic transmission of intestinal pathogens through faecal contamination of bodies of water [5]. The soil might also play a major role in transmission of enteric diseases, but this crucial link to infections has not yet been well studied [6].
- Frequent foodborne and waterborne outbreaks of infectious diarrhoea have encouraged microbiologists and epidemiologists to conduct ecological studies to understand the zoonotic and anthroponotic transmission of various diarrhoeal pathogens. In community settings in India, diarrhoeal syndromes are characterized by high faecal shedding, infectivity, growth, persistence, exposure to site-specific environmental conditions, pathogen detection, and faecal contamination [7]. However, only limited information exists regarding the association between domestic exposure and zoonotic transmission of genetically diverse diarrhoeal pathogens in the eastern part of India. A pilot survey was therefore conducted in an eastern coastal province in India in order to investigate the prevalence and possible genetic diversity of major diarrhoeal agents in faecal samples from symptomatic humans, domestic animals, and soil samples from the same locality through microbiological investigations.
INTRODUCTION
- Study design and sample collection
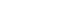
- The present cross-sectional observational study was conducted in and around the city of Bhubaneswar, located at 20.27°N 85.84°E (Figure 1) in the state of Odisha, India. Human/animal faecal samples and soil samples were collected over 13 months (March 2016 to April 2017) [8]. Samples were collected from the Khurdha district, and humans and animals of all ages were considered for this study. In Odisha, open defecation is still practiced in rural areas and even in poor and urban slum communities in urban areas such as Bhubaneswar. The most preferred site for open defecation is near ponds or paddy fields. However, people in the community and animals frequently use pond water for bathing, drinking, and other recreational activities, and farmers similarly often visit paddy fields for irrigation and other farming practices. Consequently, these sites are the highest-potential areas for anthroponotic or zoonotic transmission of enteric pathogens. Therefore, we collected samples from ponds and paddy fields.
- In total, 310 diarrhoeal human faecal samples were collected from 3 local hospitals and 2 local community clinics located a wide distance apart from each other across the study territory.
- Fresh faecal samples (n = 150) from symptomatic domestic animals (cattle, sheep, and goats) were collected across the study region. To minimize the risk of environmental contamination, fresh faeces was carefully collected from the surface of the mass that had no direct contact with the soil.
- Forty soil samples from paddy fields and banks of ponds were obtained. After collection, the samples were placed in appropriate boxes with ice packs and transported to the laboratory within 4 hours of collection. The samples were stored temporarily in a refrigerator at 4°C, and each sample was processed within 24 hours of collection.
- Immunochromatographic test
- Faecal specimens collected from symptomatic human and animal subjects were screened for rotavirus and adenovirus by an immunochromatographic test (Combi-Strip C-1004; Coris Bioconcept, Gembioux, Belgium) following the manufacturer’s instructions.
- Genomic DNA extraction and quantification
- Total faecal genomic DNA from human and animal faeces was extracted from the stool using the QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany) and soil DNA was extracted using MP Biomedicals FastDNA SPIN Kit for Soil (MP Biomedicals, Burlingame, CA, USA) following the manufacturer’s instructions.
- Polymerase chain reaction amplification and sequence analysis
- Polymerase chain reaction (PCR)-based detection was employed for various possible microbial agents. Separate primer sets were used for target-specific amplification of each microbial agent, as presented in Supplementary Material 1. For each pathogen, genomic DNA extracted from stool (Qiagen Stool DNA kit) was used as a template for PCR amplification. Genomic DNA extracted from pure culture of each microbial agent was used as a positive control in PCR screening. The PCR cycling conditions for the targeted bacterial, viral, and protozoan diarrhoeal agents were as follows: initial denaturation at 95°C for 5 minutes, followed by 34 cycles of denaturation of 94°C for 30 seconds, annealing at a primer-specific temperature at 30-45 seconds, extension at 72°C for 1 minute, and final extension for 72°C for 7 minutes. All the PCR assays were equally sensitive and specific across all different sample types, and we used the previously validated primer sets presented in Supplementary Material 1. All PCR products were subjected to 1.0-1.5% agarose gel electrophoresis to confirm the positive samples. All PCR-positive products were purified and sequenced.
- Phylogenetic analysis
- The sequences obtained from this study ([MF329642], [MF329643], [MF329644], [MF329645] [MF329646], [MF329647], [MF329648], [MF329649], [MF329650], [MF329651], [MF329652], [MF329653], [MF329654], [MF329655], [MF329656], [MF329657], [MF329658], [MF329659], [MF329660], [MF329661], [MF329662], [MF329663], [MF329664], [MF329665], [MF329666], [MF329667], [MF329668], [MF329669], [MF443209], [MF443210], [MF443211], [MF443212], [MF443213], [MF443214], [MF443215], and [MF443216]) and a few reference sequences from GeneBank ([HQ324789.1], [JQ407725.1], [AB630325.1], [JQ407711.1], [EU867486.1], [HM588724], [FR849543], [KT326927], [KY243935], [KX909565], [KU201272], [LT717486], [KP116114], [Z47381], [KP116115], [KP116113], [KP116116], [EU032322], [KF679722], [AY204229], [AY204227], [L16997], [AF159110], [JN812214], [KM199753], [AB441688], and [KM199745]) were compared for genetic relatedness. A neighbour-joining algorithm was implemented to construct a phylogenetic tree using Molecular Evolutionary Genetics Analysis version 6.0 [9].
- Statistical analysis
- On the basis of descriptive statistics, odds ratios (ORs), 95% confidence intervals (CIs), and p-values were calculated to estimate significance. The chi-square statistic was calculated using a 2 × 2 contingency table in MedCalc (MedCalc, Osted, Belgium). Principal component analysis was done using METAGENassiat [10] to analyse the possible clustering patterns of human infections acquired from animal and environmental sources.
- Ethics statement
- The study protocol was reviewed and approved by the Institutional Ethical Committee of the Kalinga Institute of Medical Sciences. Informed consent and patient datasheets were maintained for all human participant.
MATERIALS AND METHODS
Human sampling
Animal sampling
Soil sampling
- A total of 152 of 310 (49.0%) human samples, 67 of 150 (44.7%) animal samples, and 16 of 40 (40.0%) soil samples were found to be positive for at least 1 diarrhoeal pathogen. In the animals, the overall diarrhoeal pathogen detection rate was highest in sheep (41.1%), followed by goats (35.5%) and cattle (33.3%). Diarrhoeagenic Escherichia coli (DEC) was the major enteric pathogen detected in humans (28.7%), animals (38.7%), and soil (32.5%) (Table 1).
- In humans, rotavirus was detected in 17.4% of cases, followed by enteropathogenic E. coli (EPEC) (15.5%) Shigella (13.9%), Shiga toxin-producing E. coli (STEC) (9.7%), enterohemorrhagic E. coli (EHEC) and enteroaggregative E. coli (EAEC) (4.5%), Cryptosporidium and adenovirus (3.9%), and Giardia (0.6%) (Table 1). In animals, STEC (28.0%), EPEC (14.7%), and EHEC (14.0%) were the major types of DEC detected, followed by Cryptosporidium (10.0%), adenovirus (4.7%), Shigella (3.3%), and Giardia (0.7%) (Table 1). In the samples, EPEC (30.0%) was the major enteric pathogen detected in soil samples, followed by Shigella (25.0%), STEC (15.0%), Giardia (7.5%) and Cryptosporidium (5.0%) (Table 1).
- Sheep were found to be slightly more infected with DEC (41.7%) than goats (35.5%) and cattle (33.3%) (Table 2). Cryptosporidium were more often observed in goats (17.8%), while Shigella infection was predominant in sheep (6.7%). However, cattle were more likely to be positive for adenovirus than sheep and goats (Table 2). The distributions of diarrhoeal pathogens by age in humans and animals are shown in Table 3.
- In our study, we observed coinfections with different combinations of bacterial, viral, and protozoan pathogens in both faecal and soil samples. Pathogens were detected simultaneously in 39.5% of human samples, 61.2% of animal samples, and 81.2% of soil samples (Table 4). Multiple pathogens were detected significantly more frequently in soil samples (p=0.009), followed by human samples (p=0.003) and animal samples (p=0.030). In humans, Shigella and STEC (8.5%) was the most common coinfection followed by rotavirus and EPEC (7.2%) and Shigella and EPEC (6.6%) (Table 4). The most frequent combination in animals was EHEC and STEC (23.4%) (Table 4). From all positive animal samples, we observed the highest percentage of coinfections in sheep (70.8%), followed by cattle (50.0%) and goats (37.5%). The combinations of Shigella and EPEC (37.5%) and STEC and EPEC (31.2%) were predominant in soil samples (Table 4).
- Phylogenetic analysis was done to investigate the genetic relatedness and evolutionary dynamics of the strains circulating between humans and animals in the study region. Phylogenetic trees were constructed separately for each group of pathogens (Supplementary Materials 2-5). Among the DEC strains, STEC, EPEC, EHEC, and EAEC clustered in individual nodes isolated from humans and animals were found close to each other (Supplementary Material 2). Similar patterns were observed in Shigella isolates from humans and animals (Supplementary Material 3). Cryptosporidium and adenovirus isolates showed close relatedness with other strains that were isolated from domestic animals, birds, or environmental samples (Supplementary Materials 4 and 5).
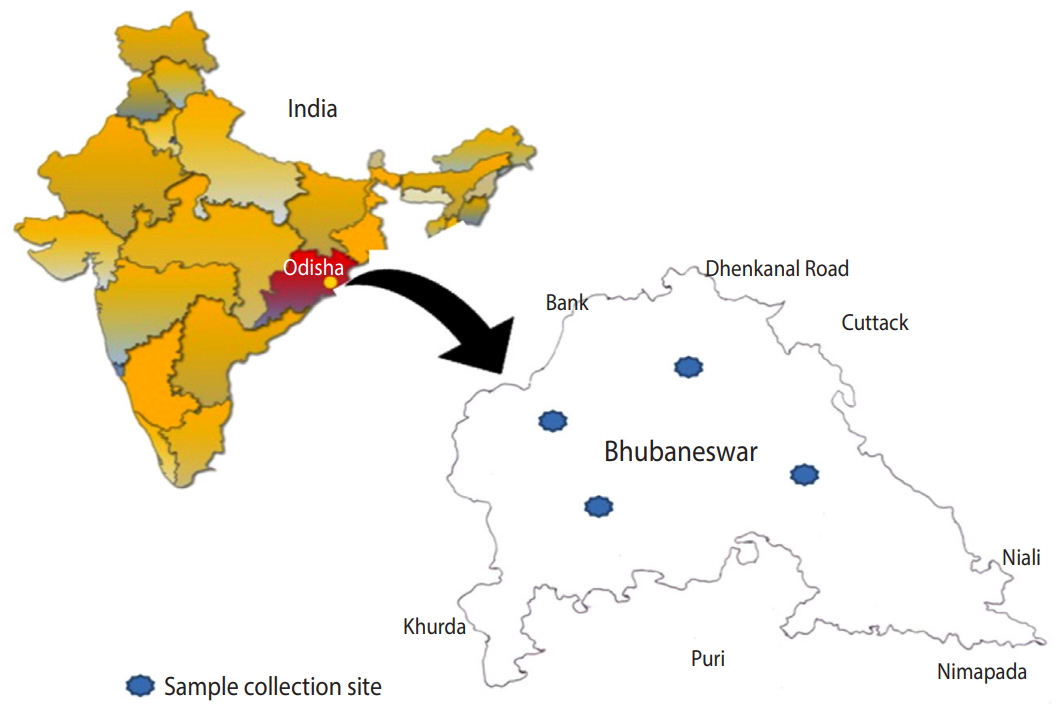
- Based on principal component analysis, 3 different clusters were generated for the human, animal, and soil samples representing the patterns of infectious agents at the genus level in the study region (Figure 2). All 3 groups shared a large portion of genera, revealing that the distribution of infections in human, animal, and soil samples was comparable. Samples collected from 4 different zones showed very similar patterns of distribution of infectious agents. All detected pathogens were distributed throughout the study area.
RESULTS
- Many resource-poor or developing countries have limited sanitary infrastructure, accompanied by a lack of awareness among communities that are suspected to be deprived of adequate education and awareness. Irrespective of focal urbanization and development of sanitation facilities, communities in underdeveloped pockets in and around urban areas are commonly reported to have higher rates of infections, particularly those associated with enteric pathogens [11]. This also places urban populations at a higher risk of acquiring infections because of their dependence on the communities living in the outskirts. In the present study, at least 1 diarrhoeal agent was detected in 40–50% of environmental samples and samples from animals and humans. When comparing the isolates from these 3 sources, we observed genetic similarities among the isolates, indicating the possibility of circulation of these microbial agents among humans, animals, and the environment in the study region.
- Pathogenic E. coli was present in 32.5% of the soil samples. The detection of pathogenic E. coli in soil was also reported in another recent study from Kenya [12]. Hence, it is important to evaluate the environmental sources (soil and water) that might play an important role in retaining diarrhoeal pathogenic agents and act as a source of infection transmission, affecting both humans and animals.
- Cattle and other ruminant animals might serve as reservoirs of STEC strains that are potentially pathogenic in humans [13,14]. In our study, STEC was detected most frequently in cattle (26.7%), followed by soil (15.0%) and human samples (9.7%). Similar patterns were observed in a previous study from Tanzania, although the prevalence rates were lower (cattle, 9.0%; humans, 3.2%; and soil, 0.8%) [15].
- Open defecation by animals and humans is a major contributor to microbial shedding into the environment (soil and water). Thus, various microbial agents from the soil can contaminate nearby community bodies of water, thereby exposing both humans and animals. Therefore, the soil can be a potential mode of transmission of diarrhoeal pathogens in low-income countries; Pickering et al. [16] were able to isolate pathogenic E. coli, enterovirus, rotaviruses, and human Bacteriodales from soil samples. In our study, we detected Shigella, EPEC, STEC, Cryptosporidium, and Giardia isolates in soil samples from the locality, while animal and human diarrhoeal cases were also found to harbour similar aetiological agents. Sequencing of the human EPEC and EHEC virulence genes eaeA and aggR showed similarities with the corresponding animal isolates. In contrast, the Cryptosporidium 18s rRNA sequence and adenovirus hexon gene sequence were similar to other Cryptosporidium parvum and adenovirus strains, respectively, that were isolated from domestic animals, birds, or environmental samples [17,18].
- Molecular epidemiological studies of pathogenic E. coli have suggested that cattle, sheep, and goats are potential sources of diarrhoeagenic EPEC, EHEC, and STEC [5,19-21]. In our study, the frequency of detection of STEC, EPEC, and EHEC was higher in sheep than in goats and cattle. Similar results were reported in another study from Turkey, where the isolation rate of STEC, EPEC, and EHEC was higher in sheep and goats [22]. STEC have been found to be closely related genetically when isolates from cattle [23] and sheep [24] were compared. Our study showed the presence of STEC in both humans and animals, suggesting possible zoonotic transmission of this pathogenic strain of E. coli.
- According to the results of the present study, Shigella was the third most common aetiological agent detected in symptomatic humans and animals in the study region. Cryptosporidium and Giardia were the other major enteric pathogens detected in all 3 sources, while adenovirus was detected in humans and animals only. Odagiri et al. [25] reported that adenovirus, Giardia, and Cryptosporidium were found in rural India with a higher prevalence than in our study.
- In comparison to a single pathogen, the presence of multiple diarrhoeal pathogens might cause more severe diarrhoea and disease pathogenesis [26]. In one of our previous studies, we detected multiple diarrhoeal pathogens, similar to the findings of other studies [27]. In the present study, multiple diarrhoeal pathogens were detected significantly more often in humans, animals, and soil samples than single pathogens. The most common pairs of concurrent pathogens in this study were Shigella and STEC in humans, EHEC and STEC in animals, and STEC and EPEC in soil samples. Given the paucity of data on the rates of specific coinfections with multiple diarrhoeal pathogens in human, animal, and soil samples, it is difficult to say whether our data are within the expected range. This is certainly an area that needs further investigation to obtain a better understanding of patterns of coinfection and their associations with disease transmission dynamics.
- In order to understand the genetic relatedness between the isolated pathogenic strains, sequencing was carried out and a phylogenetic tree was constructed. The phylogenetic tree showed similarities between the human, animal, and soil isolates. We observed that few DEC strains, such as EPEC, EAEC and EHEC, and Shigella shared similar genetic sequences and clustered under the same branch. This suggests the possible zoonotic transmission of DEC and Shigella between humans and domesticated animals in the study area. A phylogenetic tree analysis of Cryptosporidium and adenovirus found that these isolates showed sequence similarities with previously isolated human and animal strains.
- Livestock is an important reservoir for a number of enteric pathogens that can affect human and animal health. A recent study suggested that 15 major enteric pathogens are responsible for zoonotic transmission in low-income and middle-income countries (LMICs), of which 5 enteric pathogens cause approximately 1 million annual deaths [28]. Systematic reviews have demonstrated that after the introduction of improved sanitation in LMICs, a 30-40% decrease in childhood diarrhoea occurred [29,30]. Interventional sanitation efforts may reduce the quantity of human excreta in the environment, but animals are still often present in the domestic environment in LMICs, and people in these countries may have frequent contact with them [31-33]. Thus, contamination from animal faeces may still contribute to a substantial burden of disease in humans. This study demonstrated the coexistence of potential diarrhoeal enteric pathogens in human, animal, and soil samples in the study region, suggesting the possibility of zoonotic and anthroponotic transmission.
- Successive federal government programmes have emphasised building toilets to end open defecation. The current programme, the Swachh Bharat Mission, aims to provide sanitation to all households to end open defecation by October 2019. Prior to the launch of the cleanliness campaign, the coverage of sanitation in the state of Odisha was a mere 10.9%, and after all the efforts of the last 3 years sanitation coverage reached upto 70%, Odisha remains amongst the lowest-performing states, with nearly one-third of the population still not having access to toilets [34,35]. The present study area included many underdeveloped pockets in and around the city of Bhubaneswar, where access to improved sanitation is poor. This results in the practice of open defecation by large segments of the population. Direct dispersal of animal excreta into the environment is also common throughout the study region. This increases the potential risk of transmission of faecal pathogens in exposed communities. The present findings provide preliminary evidence of the diversity of potential possible transmission patterns of bacterial, viral, and protozoal diarrhoeal pathogens and provide an improved understanding of the distribution of these pathogens in humans, animals, and the shared environment (soil). Overall, this study will be helpful for expanding our knowledge of disease transmission in this region, so that transmissible diseases of concern can be controlled, thereby enhancing quality of life for the community.
- Although the present observational study deployed a unique approach to study both animal and human pathogens from the same locality, there are still some limitations, particularly in the choice of environmental samples and the sample numbers. As discussed previously, only soil samples near ponds or paddy fields were investigated based on the assumption that these are the most preferred sites for open defecation, as locations where both human and animals visit frequently for various daily activities. The initial plan to include similar numbers of human/animal faecal and soil samples could not be fulfilled due to various reasons, including but not limited to funding, time constraints, and the exclusion of a few samples due to unavoidable technical errors during transportation from the field to the analysing laboratory. To better explore the role of zoonotic transmission, pairing of human faecal, animal faecal, and soil samples collected from the same locality would be valuable, as would information on animal ownership and contact with livestock; however, these factors went beyond the scope of this study. Therefore, larger and longitudinal cohort studies of infants, children, adults, animals, soil, and community water sources would provide improved estimates of the prevalence of these diarrhoeal pathogens and their transmission in the community.
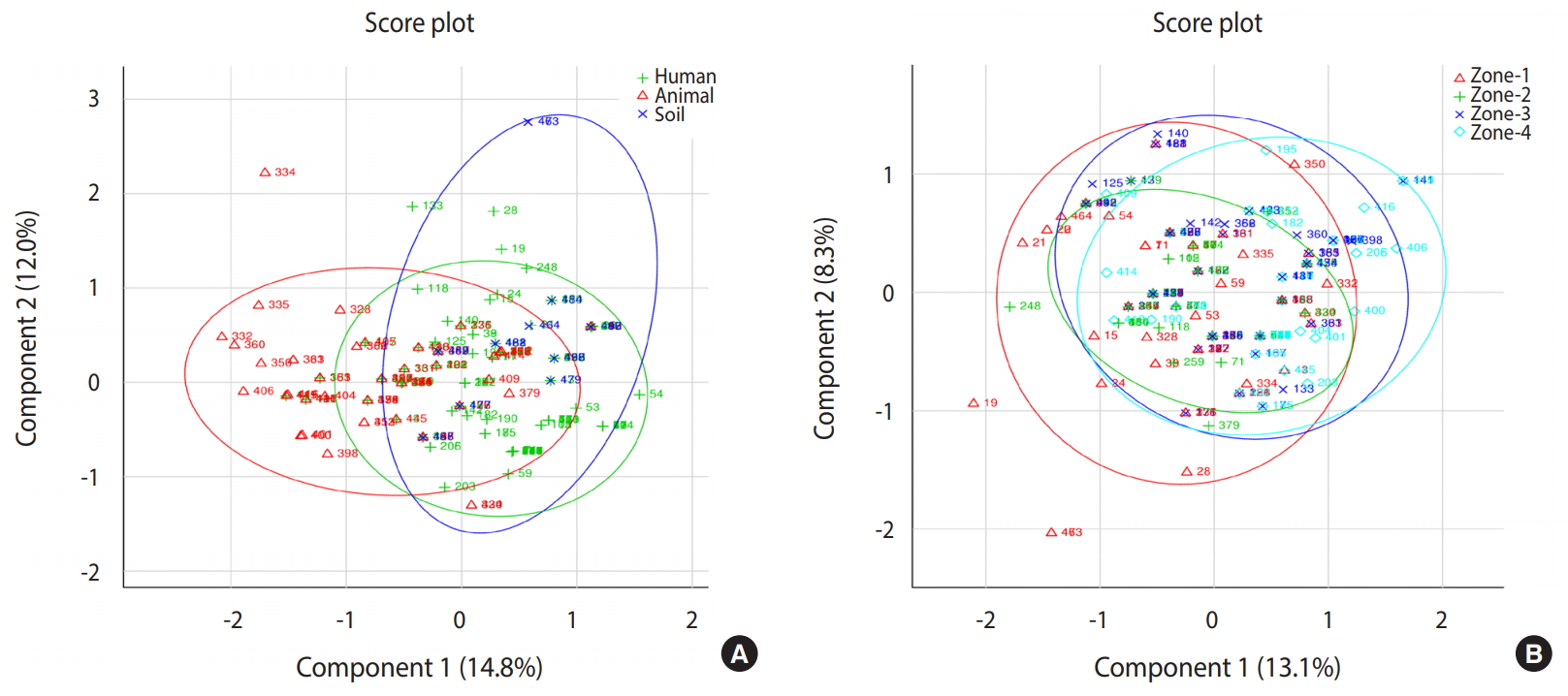
- The present study provides an improved understanding of the distribution of major enteric pathogens coexisting in humans and animals in the region, thereby suggesting a high potential for transmission among livestock and communities residing in the studied locality via contaminated soil and/or water (Figure 3). Future research on zoonotic and anthroponotic transmission of faecal contaminants should involve host-specific markers to determine the precise pathways of pathogen transmission in the region.
DISCUSSION
Electronic Supplementary Material
-
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare for this study.
-
FUNDING
This study was supported primarily by institutional funding as provided by the KIIT University, Bhubaneswar (India). The Cryptosporidium and Giardia diagnostic reagents were used from the fund granted by University of California, Davis via the London School of Hygiene and Tropical Medicine under the Orissa Rural Sanitation Health Impact Trial (No. NCT01214785).
-
AUTHOR CONTRIBUTIONS
Conceptualization: AKS. Data curation: AKS, NKM, SDP. Formal analysis: AKS, SK. Funding acquisition: PSS. Methodology: AKS, PSS. Project administration: AKS, NKM. Visualization: PSS, SK. Writing – original draft: AKS. Writing – review & editing: PSS, AKS, SP, NKM, SDP, SK.
NOTES
ACKNOWLEDGEMENTS
| Infectious agent |
Humans (n=310) |
Animals (n=150) |
Soil (n=40) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Positive, n (%) | OR | p-value | Positive, n (%) | OR | p-value | Positive, n (%) | OR | p-value | |
| DEC1 | 89 (28.7) | 62.01 | <0.001 | 58 (38.7) | 46.96 | <0.001 | 13 (32.5) | 9.14 | 0.005 |
| STEC | 30 (9.7) | 16.49 | <0.001 | 42 (28.0) | 4.08 | 0.210 | 6 (15.0) | 15.26 | 0.060 |
| EPEC | 48 (15.5) | 28.19 | <0.001 | 22 (14.7) | 25.60 | 0.001 | 12(30.0) | 35.52 | 0.010 |
| EHEC | 14 (4.5) | 7.28 | 0.009 | 21 (14.0) | 2.01 | 0.560 | 0 (0.0) | NA | NA |
| EAEC | 14 (4.5) | 7.28 | 0.009 | 4 (2.7) | 4.08 | 0.210 | 0 (0.0) | NA | NA |
| O157 | 10 (3.2) | 5.13 | 0.030 | 7 (4.7) | 7.29 | 0.060 | 3 (7.5) | 7.56 | 0.180 |
| Shigella | 43 (13.9) | 24.78 | <0.001 | 5 (3.3) | 5.13 | 0.130 | 10 (25.0) | 27.88 | 0.020 |
| Rotavirus | 54 (17.4) | 32.46 | <0.001 | 4 (2.7) | 4.08 | 0.210 | NA | NA | NA |
| Adenovirus | 12 (3.9) | 6.20 | 0.010 | 7 (4.7) | 7.29 | 0.060 | 0 (0.0) | NA | NA |
| Cryptosporidium | 12 (3.9) | 6.20 | 0.010 | 15 (10.0) | 16.55 | 0.006 | 2 (5.0) | 5.25 | 0.280 |
| Giardia2 | 2 (0.6) | 1.00 | - | 1 (0.7) | 1.00 | - | 3 (7.5) | 7.56 | 0.180 |
OR, odds ratio; DEC, diarrhoeagenic Escherichia coli; STEC, Shiga toxin-producing E. coli; EPEC, enteropathogenic E. coli; EHEC, enterohemorrhagic E. coli; EAEC, enteroaggregative E. coli; NA, not applicable.
1 The chi-square statistic was calculated using a 2×2 contingency table; a similar analysis was carried out previously by Daniels et al. [32].
2 For humans and animals, the Giardia samples were used as reference.
- 1. Liu L, Johnson HL, Cousens S, Perin J, Scott S, Lawn JE, et al. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet 2012;379:2151-2161.ArticlePubMed
- 2. Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 2013;382:209-222.ArticlePubMed
- 3. Clasen T, Boisson S, Routray P, Torondel B, Bell M, Cumming O, et al. Effectiveness of a rural sanitation programme on diarrhoea, soil-transmitted helminth infection, and child malnutrition in Odisha, India: a cluster-randomised trial. Lancet Glob Health 2014;2:e645-e653.ArticlePubMed
- 4. Zambrano LD, Levy K, Menezes NP, Freeman MC. Human diarrhea infections associated with domestic animal husbandry: a systematic review and meta-analysis. Trans R Soc Trop Med Hyg 2014;108:313-325.ArticlePubMedPMCPDF
- 5. Bublitz DC, Wright PC, Bodager JR, Rasambainarivo FT, Bliska JB, Gillespie TR. Epidemiology of pathogenic enterobacteria in humans, livestock, and peridomestic rodents in rural Madagascar. PLoS One 2014;9:e101456.ArticlePubMedPMC
- 6. Santamaría J, Toranzos GA. Enteric pathogens and soil: a short review. Int Microbiol 2003;6:5-9.ArticlePubMedPDF
- 7. Julian TR. Environmental transmission of diarrheal pathogens in low and middle income countries. Environ Sci Process Impacts 2016;18:944-955.ArticlePubMed
- 8. von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet 2007;370:1453-1457.ArticlePubMed
- 9. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013;30:2725-2729.ArticlePubMedPMCPDF
- 10. Arndt D, Xia J, Liu Y, Zhou Y, Guo AC, Cruz JA, et al. METAGENassist: a comprehensive web server for comparative metagenomics. Nucleic Acids Res 2012;40:W88-W95.ArticlePubMedPMCPDF
- 11. Pruss-Ustun A; World Health Organization. Safer water, better health: costs, benefits and sustainability of interventions to protect and promote health; 2008 [cited 2020 Jun 4]. Available from: https://apps.who.int/iris/handle/10665/43840.
- 12. Bauza V, Ocharo RM, Nguyen TH, Guest JS. Soil ingestion is associated with child diarrhea in an urban slum of Nairobi, Kenya. Am J Trop Med Hyg 2017;96:569-575.ArticlePubMedPMC
- 13. Beutin L, Geier D, Steinrück H, Zimmermann S, Scheutz F. Prevalence and some properties of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. J Clin Microbiol 1993;31:2483-2488.ArticlePubMedPMC
- 14. Mainil JG, Daube G. Verotoxigenic Escherichia coli from animals, humans and foods: who’s who? J Appl Microbiol 2005;98:1332-1344.ArticlePubMed
- 15. Wang L, Nakamura H, Kage-Nakadai E, Hara-Kudo Y, Nishikawa Y. Comparison by multilocus variable-number tandem repeat analysis and antimicrobial resistance among atypical enteropathogenic Escherichia coli strains isolated from food samples and human and animal faecal specimens. J Appl Microbiol 2017;122:268-278.ArticlePubMed
- 16. Pickering AJ, Julian TR, Marks SJ, Mattioli MC, Boehm AB, Schwab KJ, et al. Fecal contamination and diarrheal pathogens on surfaces and in soils among Tanzanian households with and without improved sanitation. Environ Sci Technol 2012;46:5736-5743.ArticlePubMed
- 17. Abreu-Acosta N, Foronda-Rodríguez P, López M, Valladares B. Occurrence of Cryptosporidium hominis in pigeons (Columba livia). Acta Parasitol 2009;54:1.ArticlePDF
- 18. Piao J, Jiang J, Xu B, Wang X, Guan Y, Wu W, et al. Simultaneous detection and identification of enteric viruses by PCR-mass assay. PLoS One 2012;7:e42251.ArticlePubMedPMC
- 19. Nishikawa Y, Zhou Z, Hase A, Ogasawara J, Kitase T, Abe N, et al. Diarrheagenic Escherichia coli isolated from stools of sporadic cases of diarrheal illness in Osaka City, Japan between 1997 and 2000: prevalence of enteroaggregative E. coli heat-stable enterotoxin 1 gene-possessing E. coli. Jpn J Infect Dis 2002;55:183-190.PubMed
- 20. Akiyama Y, Saito E, Futai H, Ogita K, Sakae H, Fukunaga M, et al. Comprehensive study of pathogenic genes distributed in Escherichia coli isolated from cattle. Food Hyg Saf Sci 2015;56:118-122.Article
- 21. Chandran A, Mazumder A. Prevalence of diarrhea-associated virulence genes and genetic diversity in Escherichia coli isolates from fecal material of various animal hosts. Appl Environ Microbiol 2013;79:7371-7380.ArticlePubMedPMC
- 22. Turkyilmaz S, Eskiizmirliler S, Tunaligil S, Bozdogan B. Identification, characterization and molecular epidemiology of Escherichia coli isolated from lamb and goat kids with diarrhoea. Acta Vet Brno 2013;82:357-362.Article
- 23. Hussein HS. Prevalence and pathogenicity of Shiga toxin-producing Escherichia coli in beef cattle and their products. J Anim Sci 2007;85(13 Suppl):E63-E72.ArticlePubMedPDF
- 24. Madzingira O. Shiga toxin-producing E. coli isolated from sheep in Namibia. J Infect Dev Ctries 2016;10:400-403.ArticlePubMed
- 25. Odagiri M, Schriewer A, Daniels ME, Wuertz S, Smith WA, Clasen T, et al. Human fecal and pathogen exposure pathways in rural Indian villages and the effect of increased latrine coverage. Water Res 2016;100:232-244.ArticlePubMedPMC
- 26. Zhang SX, Zhou YM, Xu W, Tian LG, Chen JX, Chen SH, et al. Impact of co-infections with enteric pathogens on children suffering from acute diarrhea in southwest China. Infect Dis Poverty 2016;5:64.ArticlePubMedPMC
- 27. Shrivastava AK, Kumar S, Mohakud NK, Suar M, Sahu PS. Multiple etiologies of infectious diarrhea and concurrent infections in a pediatric outpatient-based screening study in Odisha, India. Gut Pathog 2017;9:16.ArticlePubMedPMCPDF
- 28. Delahoy MJ, Wodnik B, McAliley L, Penakalapati G, Swarthout J, Freeman MC, et al. Pathogens transmitted in animal feces in low- and middle-income countries. Int J Hyg Environ Health 2018;221:661-676.ArticlePubMedPMC
- 29. Freeman MC, Garn JV, Sclar GD, Boisson S, Medlicott K, Alexander KT, et al. The impact of sanitation on infectious disease and nutritional status: a systematic review and meta-analysis. Int J Hyg Environ Health 2017;220:928-949.ArticlePubMed
- 30. Wolf J, Prüss-Ustün A, Cumming O, Bartram J, Bonjour S, Cairncross S, et al. Assessing the impact of drinking water and sanitation on diarrhoeal disease in low- and middle-income settings: systematic review and meta-regression. Trop Med Int Health 2014;19:928-942.ArticlePubMed
- 31. Dione MM, Ikumapayi UN, Saha D, Mohammed NI, Geerts S, Ieven M, et al. Clonal differences between non-typhoidal Salmonella (NTS) recovered from children and animals living in close contact in the Gambia. PLoS Negl Trop Dis 2011;5(5):e1148.ArticlePubMedPMC
- 32. Zambrano LD, Levy K, Menezes NP, Freeman MC. Human diarrhea infections associated with domestic animal husbandry: a systematic review and meta-analysis. Trans R Soc Trop Med Hyg 2014;108:313-325.ArticlePubMedPMCPDF
- 33. Daniels ME, Shrivastava A, Smith WA, Sahu P, Odagiri M, Misra PR, et al. Cryptosporidium and Giardia in humans, domestic animals, and village water sources in rural India. Am J Trop Med Hyg 2015;93:596-600.ArticlePubMedPMC
- 34. Ministry of Drinking Water and Sanitation, Government of India. National annual rural sanitation survey (NARSS) 2018-19; 2019 [cited 2020 Jun 9]. Available from: https://jalshakti-ddws.gov.in/sites/default/files/National_Report_NARSS_2018_19.pdf.
- 35. Government of Odisha. Odisha economic survey 2017-18; 2018 [cited 2020 Jun 5]. Available from: https://pc.odisha.gov.in/Download/Economic_Survey_2017-18.pdf.
REFERENCES
Figure & Data
References
Citations

- In vitro antioxidant and antidiarrheal activities of aqueous and n-hexane extracts of Cucurbita maxima seed in castor oil-induced diarrheal rats
Habibu Tijjani, Adamu Matinja, Marwanatu Yahya, Emmanuel Aondofa, Akibu Sani
Natural Resources for Human Health.2022; 2(2): 246. CrossRef

 KSE
KSE
 PubReader
PubReader ePub Link
ePub Link Cite
Cite